



Next: Fitting (GO UVFIT and
Up: Interacting with tables
Previous: Visualization (GO UVALL and
Contents
Index
Flagging (UV_FLAG, RUN UV_MFLAG, RUN UV_CLIP)
The experience shows that flagging out PdBI data inside MAPPING
rarely improve the results. This is the consequence of several factors: 1)
The on-line PdBI system quite efficiently catch possible observations
problems (e.g. phase unlock); 2) A particular care has been taken in the
computation of the relative weights so that data taken in poor observing
conditions will almost not affect the result. Finally, it is extremely
difficult to flag data based on the data itself and a 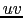 table is an
extremely compact data format which has lost most of the information needed
to make intelligent flagging. This is why we recommend the use of tools
like the ``quality assessment tool'' in CLIC which selects good
on-source data based on observing conditions (e.g. weather, tracking, ...).
This said, MAPPING offers several tools to enable data flagging in the
few cases where it can be useful like the presence of a single clearly
outlier visibility.
table is an
extremely compact data format which has lost most of the information needed
to make intelligent flagging. This is why we recommend the use of tools
like the ``quality assessment tool'' in CLIC which selects good
on-source data based on observing conditions (e.g. weather, tracking, ...).
This said, MAPPING offers several tools to enable data flagging in the
few cases where it can be useful like the presence of a single clearly
outlier visibility.
- UV_FLAG command
- Display
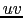 data (controled by similar
variables than GO UVALL) and calls the cursor to interactively
select a region where
data (controled by similar
variables than GO UVALL) and calls the cursor to interactively
select a region where 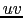 data will be flagged. The data are flagged by
changing the sign of the weight. Subsequent mapping with UV_MAP
will ignore flagged data. The flagged UV table can be saved on a file
with command WRITE UV File. The /RESET option can be
used to unflag the data (all the flagged data, not interactive).
data will be flagged. The data are flagged by
changing the sign of the weight. Subsequent mapping with UV_MAP
will ignore flagged data. The flagged UV table can be saved on a file
with command WRITE UV File. The /RESET option can be
used to unflag the data (all the flagged data, not interactive).
- UV_FLAG task
- Flag or unflag a specified time range in a
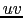 data set, for one or all baselines and a specified channel range. HOW CAN WE SEE EASILY THE TIME ?
data set, for one or all baselines and a specified channel range. HOW CAN WE SEE EASILY THE TIME ?
- UV_CLIP task
- Clip
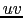 data with amplitudes higher than a
given threshold. For spectroscopic data, all channels are flagged if any
channel between the first and last considered has an amplitude greater
than the trheshold. Clipped channels are set to zero amplitude and zero
weight. It doesn't seem to work. First due to old data format,
but after using uvt_convert it does not work neither
data with amplitudes higher than a
given threshold. For spectroscopic data, all channels are flagged if any
channel between the first and last considered has an amplitude greater
than the trheshold. Clipped channels are set to zero amplitude and zero
weight. It doesn't seem to work. First due to old data format,
but after using uvt_convert it does not work neither
- UV_MFLAG task
- Compare the PdBI uv table with an uv table
calculated from a model source and flag the data if the REAL part of the
visibility differs from the model in more that a given tolerance. Data
are flagged setting weights to 0. NOT TESTED




Next: Fitting (GO UVFIT and
Up: Interacting with tables
Previous: Visualization (GO UVALL and
Contents
Index
Gildas manager
2014-07-01
![]() table is an
extremely compact data format which has lost most of the information needed
to make intelligent flagging. This is why we recommend the use of tools
like the ``quality assessment tool'' in CLIC which selects good
on-source data based on observing conditions (e.g. weather, tracking, ...).
This said, MAPPING offers several tools to enable data flagging in the
few cases where it can be useful like the presence of a single clearly
outlier visibility.
table is an
extremely compact data format which has lost most of the information needed
to make intelligent flagging. This is why we recommend the use of tools
like the ``quality assessment tool'' in CLIC which selects good
on-source data based on observing conditions (e.g. weather, tracking, ...).
This said, MAPPING offers several tools to enable data flagging in the
few cases where it can be useful like the presence of a single clearly
outlier visibility.